-Search query
-Search result
Showing 1 - 50 of 155 items for (author: kuang & x)

EMDB-43753: 
Yeast U1 snRNP with humanized U1C Zinc-Finger domain
Method: single particle / : Shi SS, Kuang ZL, Zhao R

PDB-8w2o: 
Yeast U1 snRNP with humanized U1C Zinc-Finger domain
Method: single particle / : Shi SS, Kuang ZL, Zhao R

EMDB-36678: 
PSI-AcpPCI supercomplex from Amphidinium carterae
Method: single particle / : Li ZH, Li XY, Wang WD

EMDB-36742: 
PSI-AcpPCI supercomplex from Symbiodinium
Method: single particle / : Li ZH, Li XY, Wang WD

EMDB-36743: 
PSI-AcpPCI supercomplex from Symbiodinium
Method: single particle / : Li XY, Li ZH, Wang WD

PDB-8jw0: 
PSI-AcpPCI supercomplex from Amphidinium carterae
Method: single particle / : Li ZH, Li XY, Wang WD

PDB-8jze: 
PSI-AcpPCI supercomplex from Symbiodinium
Method: single particle / : Li ZH, Li XY, Wang WD

PDB-8jzf: 
PSI-AcpPCI supercomplex from Symbiodinium
Method: single particle / : Li XY, Li ZH, Wang WD

EMDB-35987: 
Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana
Method: single particle / : Shen LL, Li ZH, Shen JR, Wang WD

EMDB-36054: 
Structure of FCP trimer in Cyclotella meneghiniana
Method: single particle / : Shen LL, Li ZH, Shen JR, Wang WD

EMDB-37267: 
Structure of PSII-FCPII-G/H complex in the PSII-FCPII supercomplex from Cyclotella meneghiniana
Method: single particle / : Shen LL, Li ZH, Shen JR, Wang WD

EMDB-37268: 
Structure of PSII-FCPII-I/J/K complex in the PSII-FCPII supercomplex from Cyclotella meneghiniana
Method: single particle / : Shen LL, Li ZH, Shen JR, Wang WD

PDB-8j5k: 
Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana
Method: single particle / : Shen LL, Li ZH, Shen JR, Wang WD

PDB-8j7z: 
Structure of FCP trimer in Cyclotella meneghiniana
Method: single particle / : Shen LL, Li ZH, Shen JR, Wang WD

PDB-8w4o: 
Structure of PSII-FCPII-G/H complex in the PSII-FCPII supercomplex from Cyclotella meneghiniana
Method: single particle / : Shen LL, Li ZH, Shen JR, Wang WD

PDB-8w4p: 
Structure of PSII-FCPII-I/J/K complex in the PSII-FCPII supercomplex from Cyclotella meneghiniana
Method: single particle / : Shen LL, Li ZH, Shen JR, Wang WD

EMDB-41097: 
cryoEM structure of Smc5/6 5mer
Method: single particle / : Yu Y, Patel DJ
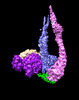
EMDB-41098: 
Smc5/6 8mer
Method: single particle / : Yu Y, Patel DJ

EMDB-35766: 
Structure and characteristics of a photosystem II supercomplex containing monomeric LHCX and dimeric FCPII antennae from the diatom Thalassiosira pseudonana
Method: single particle / : Feng Y, Li ZH, Wang WD, Shen JR

EMDB-35899: 
FCP heterodimer, Lhca2, and Lhcf5 together as the M1 side binds to the PSII core in the diatom Thalassiosira pseudonana
Method: single particle / : Li Z, Feng Y, Wang W, Shen JR

PDB-8iwh: 
Structure and characteristics of a photosystem II supercomplex containing monomeric LHCX and dimeric FCPII antennae from the diatom Thalassiosira pseudonana
Method: single particle / : Feng Y, Li ZH, Wang WD, Shen JR

PDB-8j0d: 
FCP heterodimer, Lhca2, and Lhcf5 together as the M1 side binds to the PSII core in the diatom Thalassiosira pseudonana
Method: single particle / : Li Z, Feng Y, Wang W, Shen JR

EMDB-34883: 
Bry-LHCII homotrimer of Bryopsis corticulans
Method: single particle / : Li ZH, Shen JR, Wang WD

EMDB-34930: 
Bry-LHCII heterotrimer of Bryopsis corticulans
Method: single particle / : Li ZH, Shen JR, Wang WD

PDB-8hlv: 
Bry-LHCII homotrimer of Bryopsis corticulans
Method: single particle / : Li ZH, Shen JR, Wang WD

PDB-8hpd: 
Bry-LHCII heterotrimer of Bryopsis corticulans
Method: single particle / : Li ZH, Shen JR, Wang WD

EMDB-29497: 
Chimeric HsGATOR1-SpGtr-SpLam complex
Method: single particle / : Tettoni SD, Egri SB, Doxsey DD, Ouch C, Chang J, Song K, Xu C, Shen K

PDB-8fw5: 
Chimeric HsGATOR1-SpGtr-SpLam complex
Method: single particle / : Tettoni SD, Egri SB, Doxsey DD, Ouch C, Chang J, Song K, Xu C, Shen K

EMDB-29428: 
SARS-CoV-2 Spike D614G variant, pH 7.4, 0.01% CHAPS, Fully Closed
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29430: 
SARS-CoV-2 Spike D614G variant, pH 7.4, 0.01% CHAPS, One RBD Open
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29431: 
SARS-CoV-2 Spike D614G variant, pH 7.4, 0.01% CHAPS, Two RBD Open
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29432: 
SARS-CoV-2 Spike D614G variant, pH 7.4, 0.01% CHAPS, Three RBD Open
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29434: 
SARS-CoV-2 Spike D614G variant, pH 7.4, 0.5% CHAPS, Fully Closed
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29435: 
SARS-CoV-2 Spike D614G variant, pH 7.4, 0.5% CHAPS, One RBD Open
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29436: 
SARS-CoV-2 Spike D614G variant, pH 7.4, 0.5% CHAPS, Two RBD Open
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29438: 
SARS-CoV-2 Spike D614G variant, pH 7.4, 0.5% CHAPS, Three RBD Open
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29444: 
SARS-CoV-2 Spike D614G variant, pH 7.4, 0.01% DDM, Fully Closed
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29445: 
SARS-CoV-2 Spike D614G variant, pH 7.4, 0.01% DDM, One RBD Open
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29446: 
SARS-CoV-2 Spike D614G variant, pH 7.4, 0.01% DDM, Two RBD Open
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29460: 
SARS-CoV-2 Spike D614G variant, pH 7.4, 0.5% DDM, Fully Closed
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29461: 
SARS-CoV-2 Spike D614G variant, pH 7.4, 0.5% DDM, One RBD Open
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29462: 
SARS-CoV-2 Spike D614G variant, pH 7.4, 0.5% DDM, Two RBD Open
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29463: 
SARS-CoV-2 Spike D614G variant, pH 7.4, 0.5% DDM, Three RBD Open
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29464: 
SARS-CoV-2 Spike D614G variant, pH 5.0, Fully Closed
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29465: 
SARS-CoV-2 Spike D614G variant, pH 5.0, One RBD Open (RBD observed)
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29466: 
SARS-CoV-2 Spike D614G variant, pH 5.0, One RBD Open (RBD scattered)
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29467: 
SARS-CoV-2 Spike D614G variant, pH 5.0, Two RBD Open (two RBD observed)
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K

EMDB-29468: 
SARS-CoV-2 Spike D614G variant, pH 5.0, Two RBD Open (one RBD observed, one RBD scattered)
Method: single particle / : Egri SB, Wang X, Diaz-Salinas M, Luban J, Dudkina N, Munro J, Shen K
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
